Fig. 1
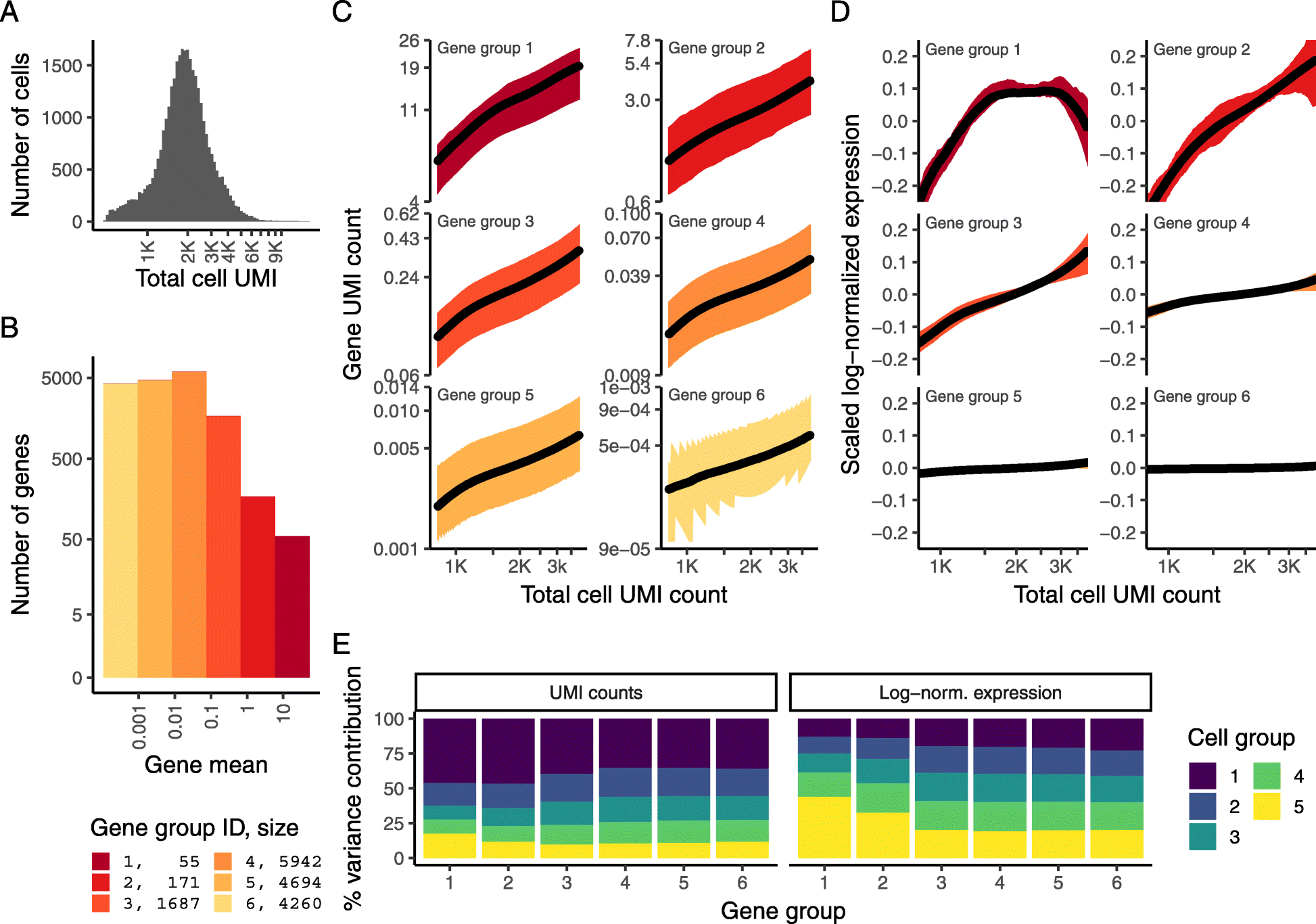
33,148 PBMC dataset from 10X Genomics. a Distribution of total UMI counts / cell (“sequencing depth”). b We placed genes into six groups, based on their average expression in the dataset. c For each gene group, we examined the average relationship between observed counts and cell sequencing depth. We fit a smooth line for each gene individually and combined results based on the groupings in b. Black line shows mean, colored region indicates interquartile range. d Same as in c, but showing scaled log-normalized values instead of UMI counts. Values were scaled (z-scored) so that a single Y-axis range could be used. e Relationship between gene variance and cell sequencing depth; cells were placed into five equal-sized groups based on total UMI counts (group 1 has the greatest depth), and we calculated the total variance of each gene group within each bin. For effectively normalized data, each cell bin should contribute 20% to the variance of each gene group
